Few factors are as crucial for the regulated course of cellular functions as the balance between structure or synthesis and the breakdown of cytoplasmic organelles and proteins. The cell regulates this balance primarily via two mechanisms: 1) the ubiquitin-proteasome system and 2) autophagocytosis (‘self-eating’), in which cellular components are fed to lysosomal degradation. While the ubiquitin-proteasome system primarily degrades short-lived and misfolded proteins, the degradation of cytoplasmic components, damaged organelles and long-lived and aggregated proteins is carried out by means of autophagocytosis. This cellular 'recycling’ system is highly regulated and thus enables the efficient return and regeneration of the cell building blocks.
Ubiquitin
Ubiquitination, one of the most common post-translational modifications (PTM) in the cell, is the mechanism by which the majority of short-lived proteins are marked for degradation and degradation. It is a highly specific system in which ubiquitin, a protein that is rather small with 76 amino acids, is linked via an amide bond to an epsilon amine of a lysine of the target protein. This is done using a three-step enzymatic process.
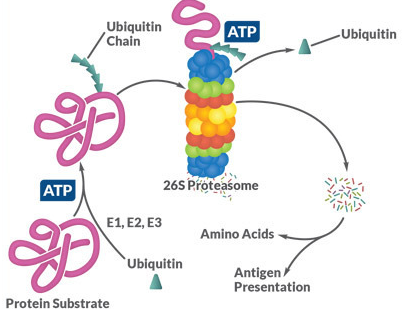
Activation of the ubiquitin by E1 (depending on ATP).
Transfer of the ubiquitin from E1 to the cysteine of the active center of the ubiquitin-conjugating (UBC) enzyme E2 (there are more than 30 different E2 proteins).
Ligation of the C-terminal glycine of the ubiquitin to the target protein by one of over 500 E3 ubiquitin ligases.
proteasome
Figure 1. The target proteins of the ubiquitin-proteasome system are ubiquitinated by E1, E2 and E3 using a three-step, ATP-dependent mechanism. The proteins are then unfolded in the proteasome and broken down into short peptide chains, which can then be used in the course of the antigen presentation via MHC I or else hydrolyzed into the individual amino acids.
As part of the hierarchical ubiquitination cascade, E1 can bind dozens of E2 proteins, which in turn have the ability to associate with hundreds of E3 proteins. Proteins can also be monoubiquitinated, multiubiquitinated and polyubiquitinated. This increases the variability of the degradation system, since different ubiquitination patterns can activate different signaling pathways by binding to more than 20 known ubiquitin binding domains. Similar to phosphorylation, ubiquitination is reversible and ∼85 deubiquitinases (DUBs) have been described. The majority of the ubiquitinated proteins are degraded, as explained above, but modification with ubiquitin can also act as a transport signal for some proteins or mediate protein-protein interactions (Figure 1) .1-3
Ubiquitination is involved in various cell processes and malfunctions, including immune response, DNA repair, signal transduction, cancer, neuropathy and many more. This naturally means that errors in the ubiquitination pathway have far-reaching and diverse effects in the affected cell and are therefore causally involved in the development of a large number of neurodegenerative, cardiovascular and tumor diseases.4,5 As part of the great chemical and technological advances (e.g. mass spectrometry) Over the past two decades, more and more active substances have therefore been developed that intervene in the ubiquitin-proteasome system.
Cayman Chemical offers you a wide range of products for researching the role and function of the ubiquitin and proteasome. An example are various inhibitors and agonists of ubiquitination, which enable the targeted analysis of individual processes or enzymes in the ubiquitin-proteasome system. In addition, Cayman offers you a range of antibodies, including FK1 and FK2, with which you can distinguish specific ubiquitination patterns on the target proteins (monoubiquitination, multiubiquitination and polyubiquitination). Cayman Chemical has also recently added kits for ubiquitination detection, quantification of ubiquitin-binding proteins, or deubiquitinase activity, which are a key tool for scientists to further explore the system.
Autophagocytosis
Autophagocytosis is an evolutionarily highly conserved cellular degradation process in which cell components such as organelles are degraded using the lysosomal machinery. As a result, autophagocytosis is a normal sub-process of cell growth, development, differentiation, and homeostasis. Autophagocytosis enables the cell to maintain a delicate balance in the synthesis, degradation and recycling of cellular components. In lower organisms, it regulates the metabolic equilibrium with fluctuating nutrient supply.6 While it was originally assumed that autophagocytosis is a rather 'crude’ and less regulated process for the destruction of cell components, it has become apparent in recent research that autophagocytosis is also 'selective’ expires. In higher organisms, this system eliminates aggregated proteins, damaged organelles such as mitochondria (mitophagy) or peroxisomes (pexophagia), and also eliminates microbial pathogens (xenophagia) (Figure 2) .7,8 Although autophagocytosis occurs permanently at least at a basic level in everyone Cell, but this activity can increase dramatically as a result of external and internal stimuli. Various signal transduction pathways convey this stringent regulation, however the exact mechanisms behind it are currently often only incompletely investigated. However, a whole range of proteins involved are known, including mTOR, p62, Beclin 1 and PINK1 (Figure 3) .8 Since autophagocytosis, like ubiquitination, must be in constant balance with the new synthesis of proteins and organelles, it can also be expected that that incorrect regulation of the system has far-reaching, pathological consequences for the organism. In fact, it has been shown that knockout mice lacking a gene essential for autophagocytosis die within hours after birth.9 It is also known that disorders in autophagocytosis play a role in cancer, neurodegenerative diseases (both in HD) and Alzheimer’s disease shows an increased accumulation of autophagic vacuoles), lysosomal storage diseases, infectious diseases and the repair of DNA damage (via HDACs). Autophagocytosis dysfunction is also thought to be involved in aging and cell death.10,11
autophagy
Figure 2. Non-selective and selective autophagocytosis (A) Non-selective autophagocytosis is activated under hunger stress and mediates the breakdown of cytoplasmic components into usable smaller building blocks. (B) Stimuli such as activation of p62, damage to mitochondria or infections can trigger the activation of selective autophagocytosis.
Autophagocytosis is primarily characterized by the presence of a unique organelle, the autophagosome. The autophagosome is a vacuole with a diameter of 300-900 nm that is surrounded by a double membrane. It arises from the so-called phagophore, which expands and includes cytoplasmic components. These phagosomes then fuse with the lysosome to form the active autolysosome. 6
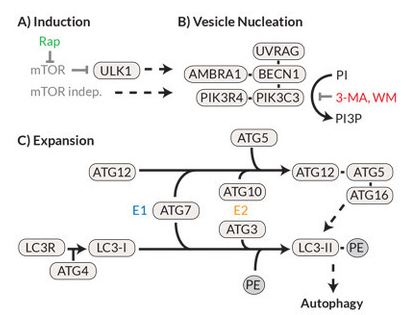
In the late 1990s, work on yeasts led to the discovery of over 20 autophagocytosis-involved genes (ATGs). Further research eventually led to the description of the mammalian homologues of these genes and the associated proteins.12, 13 A schematic representation of the signal cascade as well as the known inhibitors and activators is shown in Figure 3. Under nutrient-rich conditions, the protein kinase mTOR acts as the primary negative regulator of autophagocytosis. Under hunger conditions, however, mTOR (the rapamycin target protein in the mammal) is deactivated and thus ULK (the mammalian equivalent of the yeast protein ATG1) is activated, which then translocates into the endoplasmic reticulum and initiates the autophagocytosis cascade there. The formation of the phagosome is completed by ATG8 (in mammals the light chain 3 of the microtubule-associated protein (MAP-LC3 or LC3-I)) cleaved C-terminally by ATG4 and phosphatidylethanolamine (PE) is coupled to ATG8. This LC3 phospholipid (LC3-II) then covalently binds to the phagophore and is essential for the formation of the phagosome. For this reason, LC3-II can be used as a highly specific marker for the autophagosome. Just like ubiquitination, this process is reversible, and this delipidization is also mediated via ATG4.
Figure 3. The autophagocytosis pathway in mammals (A) The prototypical induction of autophagocytosis by mTOR (inhibited by rapamycin) leads to (B) nucleation / expansion of the autophagosomal membrane with the help of a protein complex containing Beclin 1 and PI3K (inhibited by 3-methyladenine ( 3-MA) and Wortmannin). (C) The membrane is expanded by a mechanism similar to ubiquitination, in which ATG7 and ATG3 convert LC3-I to LC3-II, which in turn accumulates on the autophagosomal membrane.
Autophagocytosis activity in cells can be quantified using many different methods. The first successful approaches included staining the autophagosomes with acidotropic dyes, such as monodansylcadaverine (MDC) (Cayman’s Autophagy / Cytotoxicity Dual Staining Kit), and transmission electron microscopy (TEM).
Interestingly, the PE modification of LC3-I causes a change in electrophoresis mobility, which enables precise analysis of the LC3-I and LC3-II levels by Western blot. In combination with lysosomal inhibitors such as bafilomycin A1, this technique can be used to measure the accumulation of LC3-II in the autophagolysosome. This in turn allows the ‘autophagic flow’ to be determined, ie the rate at which proteins and cell material are processed in the autophagosome. 14-17
The ubiquitin-proteasome system and autophagocytosis are of crucial importance for the difficult balance of the production and degradation of proteins and organelles in all cells. Today it is known that these pathways can also interact and that autophagocytosis also degrades ubiquitinated proteins under extreme stress. Although much has been deciphered, a lot of research is still needed. Researchers worldwide are working on identifying new biomarkers in order to better understand the functions of these systems and to develop innovative therapeutic options for the associated diseases.
